We are currently recruiting a postdoc and a PhD student in Quantitative Immunology!
This is a brilliant opportunity to join our very supportive and highly collaborative research group, located in one of the best cities in the world for research in quantitative immunology. We are committed to the career development of all team members, work on unique datasets and questions, and provide scope to explore and adjust projects based on candidate interests.
Beyond computational immunologists, we are particularly interested in candidates with a strong background in physics, machine learning, or maths, for which we can provide on-the-job training in immunology.
Informal inquires are very welcome (andreas.mayer@ucl.ac.uk).
EPSRC PhD project (CDT AMR): Reverse epitope discovery for Tuberculosis vaccine design using deep learning
Rationale & Background
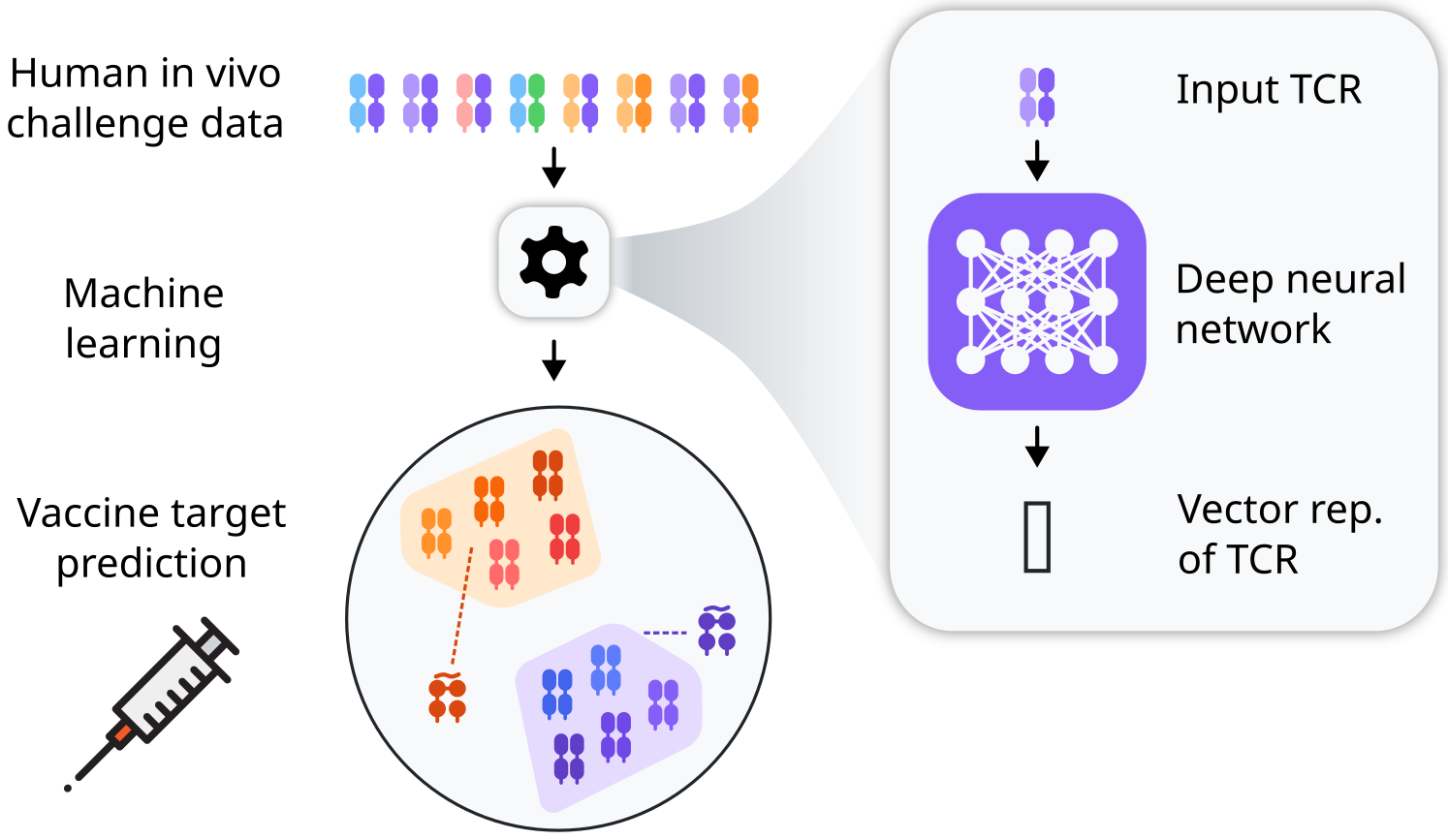 Drug-resistant tuberculosis (TB) accounts for one out of every three deaths from antimicrobial resistance. To reduce this burden more efficacious vaccines are needed. Development of better vaccines is critically limited by insufficient insights into the mechanisms of protective immunity. An emergent approach to antigen discovery starts by characterizing immunodominant T cell responses in vivo and then works back from their T cell receptor (TCR) sequence to their antigen targets [Ref. 1]. This reverse epitope discovery approach offers an exciting prospect for complex pathogens such as Mycobacterium tuberculosis (Mtb), where traditional vaccine design has had limited success. However, experimental throughput of antigen discovery remains low. In this project, we will develop deep learning approaches to prioritise peptides for experimental testing.
Drug-resistant tuberculosis (TB) accounts for one out of every three deaths from antimicrobial resistance. To reduce this burden more efficacious vaccines are needed. Development of better vaccines is critically limited by insufficient insights into the mechanisms of protective immunity. An emergent approach to antigen discovery starts by characterizing immunodominant T cell responses in vivo and then works back from their T cell receptor (TCR) sequence to their antigen targets [Ref. 1]. This reverse epitope discovery approach offers an exciting prospect for complex pathogens such as Mycobacterium tuberculosis (Mtb), where traditional vaccine design has had limited success. However, experimental throughput of antigen discovery remains low. In this project, we will develop deep learning approaches to prioritise peptides for experimental testing.
Deep learning has recently begun to achieve breakthrough success on important problems in biology, as illustrated by this year’s Nobel prize for computational protein design. However, there remain significant knowledge gaps in the application of AI to protein science. This project addresses one of the most pressing of them: Predicting protein-protein interactions between the highly variable and flexible loops of immune receptors and their ligands. Anchored by a concrete focus on developing better vaccines to TB, and driven by unique datasets and interdisciplinary collaborations, this projects aims to make transformative progress on this grand challenge in computational biology.
Aims & Objectives
- To rank putative targets by in silico predictions of MHC binding
In preliminary work, we have identified T cell receptor motifs among the TCR repertoire at the site of a Tuberculin skin test, and linked these to subject major histocompatibility complex (MHC) alleles [Ref. 2]. The binding of peptides to MHC poses a major constraint on which antigens are likely to be targeted by the TCRs of interest, which we will leverage by using existing machine learning models to probabilistically rank peptides for the MHCs of interest. - To develop deep learning approaches to prioritize targets based on TCR sequence
Recent advances allow prediction of TCR binding to pMHCs for which some experimental training data is available. We will use SCEPTR, a protein language model for TCRs developed in our group [Ref. 3], to map TCR motifs to known pMHCs. However, only few TCR-pMHC pairing are known for TB, so we will next generalise the contrastive learning approach we developed to train SCEPTR to train predictors that can rank peptides unseen during training. Progress in generalisable prediction of TCR-pMHC specificity will have substantial impact beyond TB, if successful.
Skills
This project provides a powerful platform for application of machine learning to biomedical research, invaluable to academia and industry. The student will develop expertise in large language models for the prediction of protein function, and contribute substantial new knowledge in the field. They will also acquire expertise in immunology as part of an international Wellcome Trust funded team including computational biologists, wet-lab scientists and clinicians focused on discovering the T cell determinants of protection and pathogenesis in Tuberculosis.
→ Apply by January 26th
Wellcome Trust funded Research Fellow: Clonal and functional T cell determinants of protection and pathogenesis in tuberculosis
The post is supported by a Wellcome Trust Discovery Award for identification of clonal and functional T cell determinants of protection and pathogenesis in tuberculosis, aiming to develop novel approaches to stratifying disease risk in people who become infected and inform the design of next-generation vaccines. This programme of work is underpinned by the hypothesis that protective and pathogenic immunity is determined at the level of highly specific T cell-peptide/MHC interactions mediated by generalisable T cell metaclones. We aim to identify these public metaclones by TCR sequencing and undertake reverse epitope discovery to identify their pMHC targets.
The research fellow will be responsible for innovation, refinement and implementation of the computational approaches for identification of public T cell metaclones from large scale TCR sequencing data; and to advance reverse epitope discovery by prioritizing epitopes using information from pathogen transcriptomic/proteomic datasets, and by supporting novel development of machine learning models that predict TCR-pMHC interactions.
The project involves an international consortium. The post holder will report to computational biologist Dr. Andreas Tiffeau-Mayer (andreas.mayer@ucl.ac.uk), but will also work closely with “wet” lab scientists, clinicians and computational biologists within the Innate2Adaptive group (led by Mahdad Noursadeghi, Benny Chain and Hans Stauss) and our project partners in South Africa (led by Al Leslie at the Africa Health Research Institute).
The post is for 2 years in the first instance, with the possibility of extension.
The position will suit a highly motivated postdoctoral computational scientist interested in host-pathogen interactions which determine the outcomes of infection.
Applicants should have a PhD (or about to be awarded) in a relevant subject, and experience of application of bioinformatics, computational science and mathematical modelling of biological systems. Experience of sequencing data, immunology and infectious disease research are also desirable. Good oral and written communication skills are essential, as is commitment to effective multi-disciplinary teamwork.